Part B2: How to choose which xyz files to optimise in VASP
After running PartB in Adsorber, the Adsorber program will create a number of xyz files, where each adsorbate has been bound to each top, bridge, three-fold, and four-fold binding site across your system. However, you may not want to optimise everyone of these binding sites and want to be selective.
What you want do is to choose is to select the adsorbates in the sites and orientations that you want to optimise in VASP from the Part_B_All_Systems_with_Adsorbed_Species folder and copy them into the Part_C_Selected_Systems_with_Adsorbed_Species_to_Convert_into_VASP_files folder.
In this section, we will discuss how to choose xyz files for further optimise in VASP. This section requires the use of the ASE GUI and Jmol to view our chemical systems. The installation and use of these visualisation programs is given in External Programs that will be useful to install for using Adsorber.
How to choose which xyz files to optimise in VASP
To help determine which surface sites to adsorb adsorbates to, Adsorber will create a folder called Part_B_Binding_Site_Locations that contains four xyz files that show all the binding sites found in your system. These are:
SYSTEM_NAME_top_sites.xyz: Thisxyzfile contains all the top sites across your system.SYSTEM_NAME_bridging_sites.xyz: Thisxyzfile contains all the bridging sites across your system.SYSTEM_NAME_three_fold_sites.xyz: Thisxyzfile contains all the three-fold sites across your system.SYSTEM_NAME_four_fold_sites.xyz: Thisxyzfile contains all the four-fold sites across your system.
In these systems, the binding sites are represented as hydrogen atoms. Examples of SYSTEM_NAME_top_sites.xyz, SYSTEM_NAME_bridging_sites.xyz, and SYSTEM_NAME_three_fold_sites.xyz of a Cu78 cluster are given below. This particular example does not contain any four-fold sites.
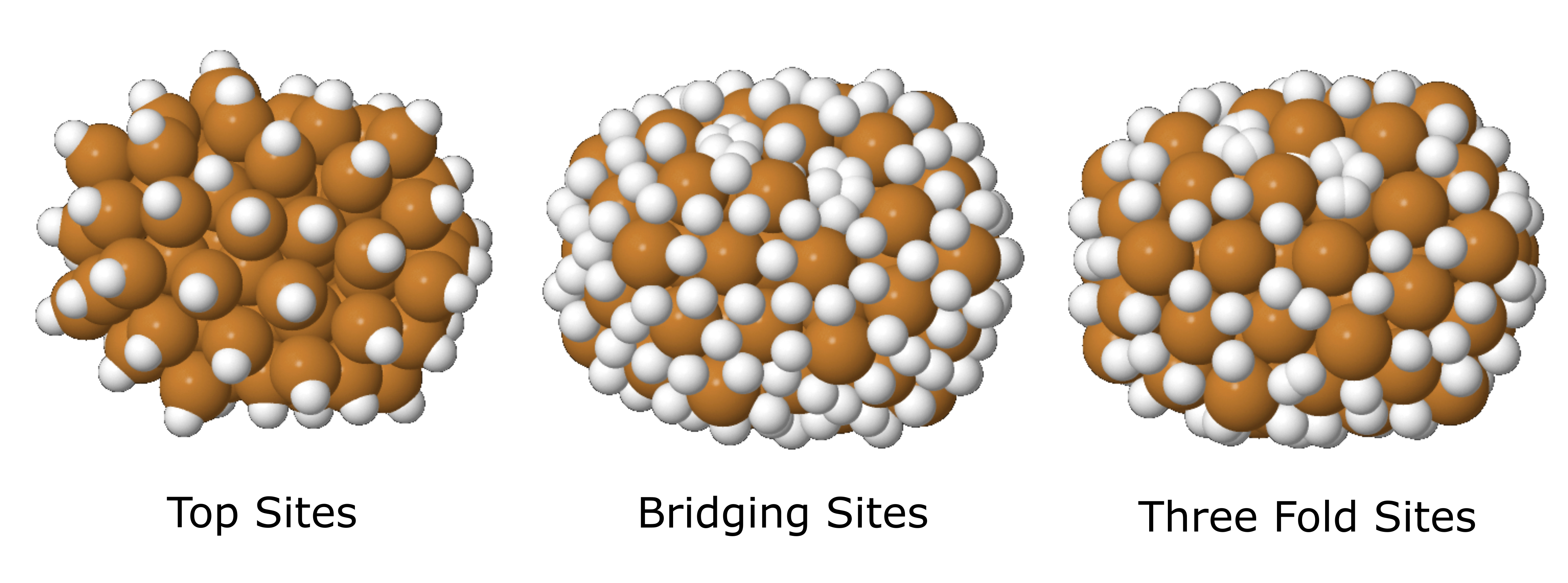
Top sites (SYSTEM_NAME_top_sites.xyz), bridging sites (SYSTEM_NAME_bridging_sites.xyz), and three-fold sites (SYSTEM_NAME_three_fold_sites.xyz) across this cluster, where each of these sites are represented with hydrogen atoms.
The xyz files that are found in Part_B_All_Systems_with_Adsorbed_Species\ADSORBATE\ADSORPTIONSITE are labelled as ADSORBATE_ADSORPTIONSITE_Label_Index.xyz, where:
ADSORBATE: The adsorbate you want to adsorb to the surface of your system.ADSORPTIONSITE: The type of surface site that the adsorbate is bound to.Label: The label of the binding site.Index: The index of the binding site.
For example, if you want to see the COOH molecule bound to three-fold site labelled 44, you would go to Part_B_All_Systems_with_Adsorbed_Species > COOH > Three_Fold_Sites and look at any of the file with COOH_three_fold_sites_44 in its name. This example is shown below, next to the original three-fold binding site .xyz file.
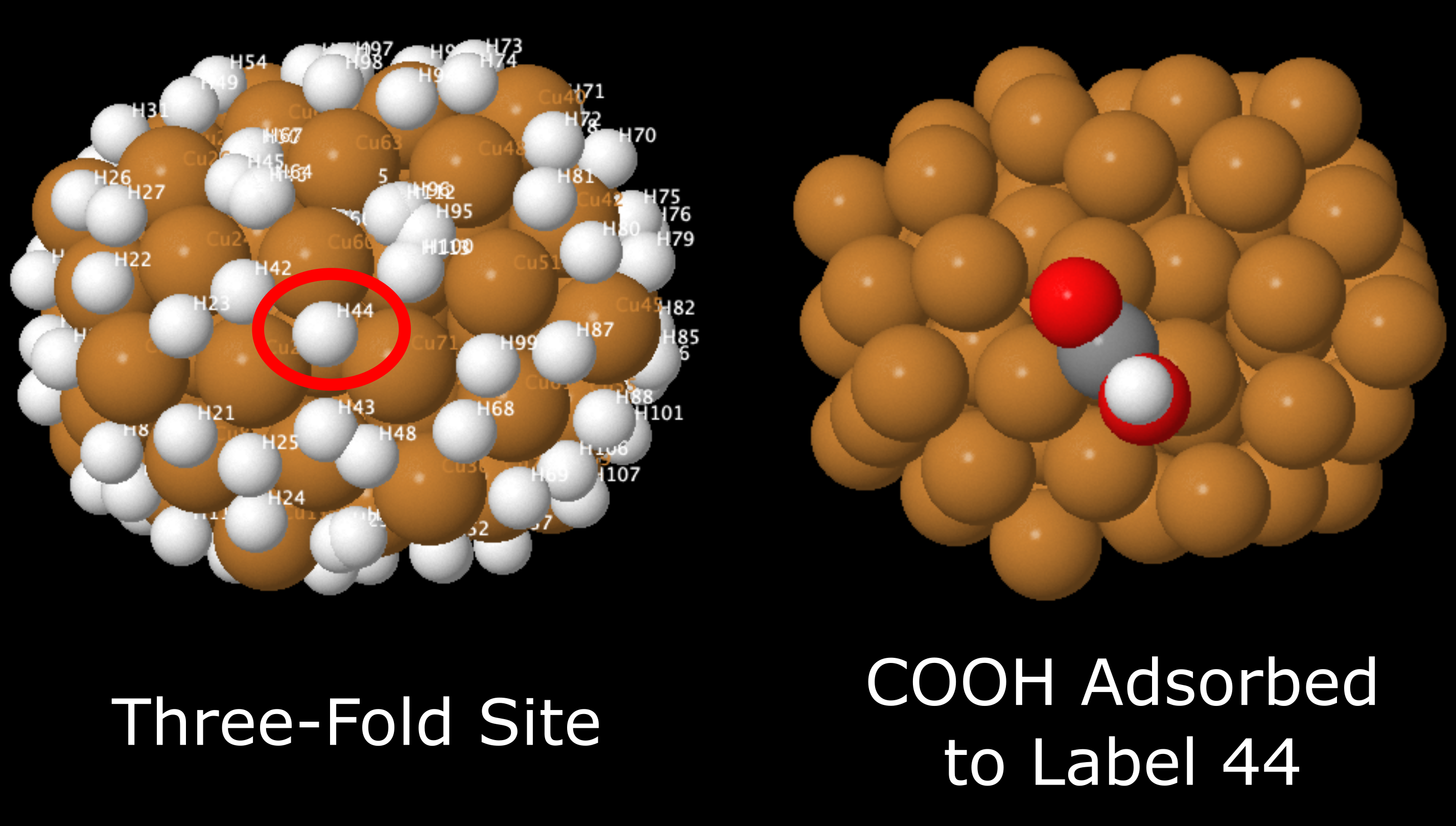
This example cluster with a COOH molecule adsorbed to three-fold site labelled 44 (just one of the orientations is shown in this example).
Selecting binding sites using the Label command in Jmol
You can view the Label of each binding site in Jmol. This is the number that is assign to each of the binding sites. To do this, first open the xyz file in the terminal:
jmol SYSTEM_NAME_top_sites.xyz
jmol SYSTEM_NAME_bridging_sites.xyz
jmol SYSTEM_NAME_three_fold_sites.xyz
jmol SYSTEM_NAME_four_fold_sites.xyz
This will open up your cluster/surface model in Jmol. Then in the Jmol menu click Display > Label > Name. This will label all the atoms by their element symbol and Label, where the binding site are labelled HX, where X is the Label of the hydrogen/binding site in the cluster/surface model.
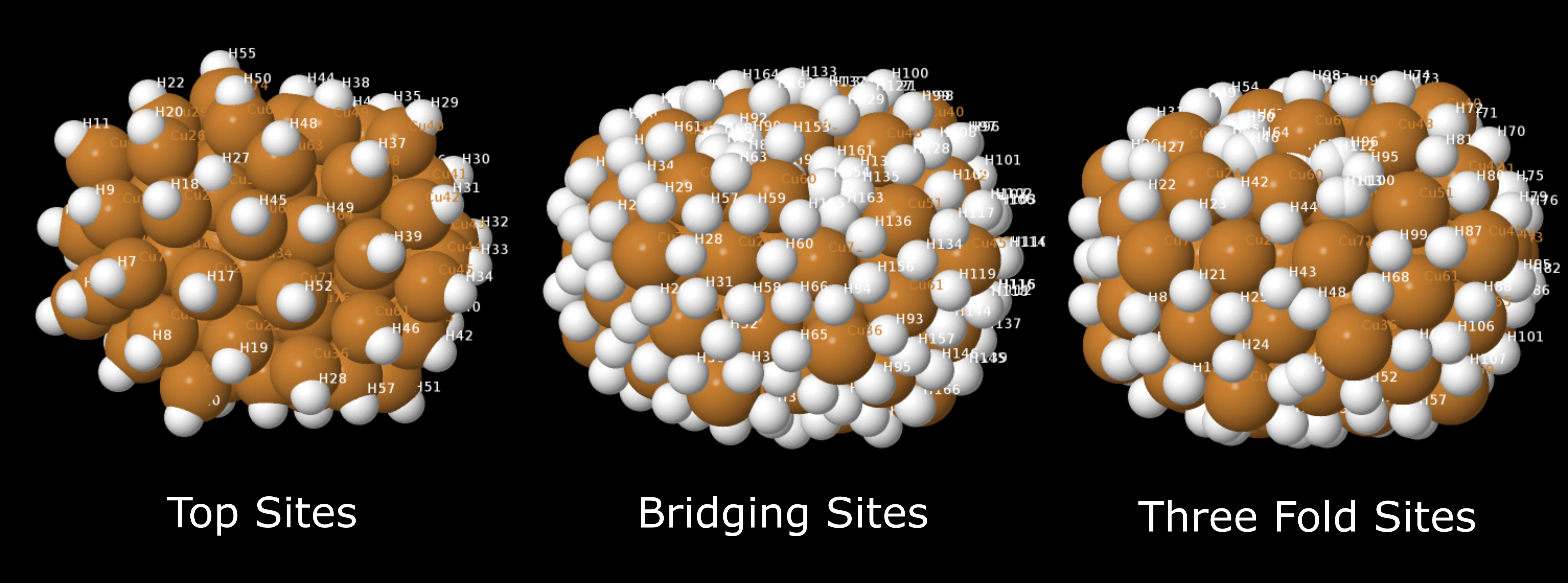
Top sites (SYSTEM_NAME_top_sites.xyz), bridging sites (SYSTEM_NAME_bridging_sites.xyz), and three-fold sites (SYSTEM_NAME_three_fold_sites.xyz) across this cluster, where each of these sites are represented with hydrogen atoms. Each site is labelled HX, where X is the Label for that binding site.
Advice on how I Choose xyz Files for VASP Optimise with Adsorber
The way that I have found the best use of these four xyz files is by colouring in the hydrogens in Jmol that I want to bind all adsorbate to on this system. This can be the same colour, or by colour in the different types of sites in different colours that are of use to use. For example, in the following figure I have coloured the binding sites of interest across this Cu78 cluster green for icosahedral sites, interesting sites about the middle of the cluster in yellow, other interesting corner sites in blue, and vacant five-fold vertex sites in red.
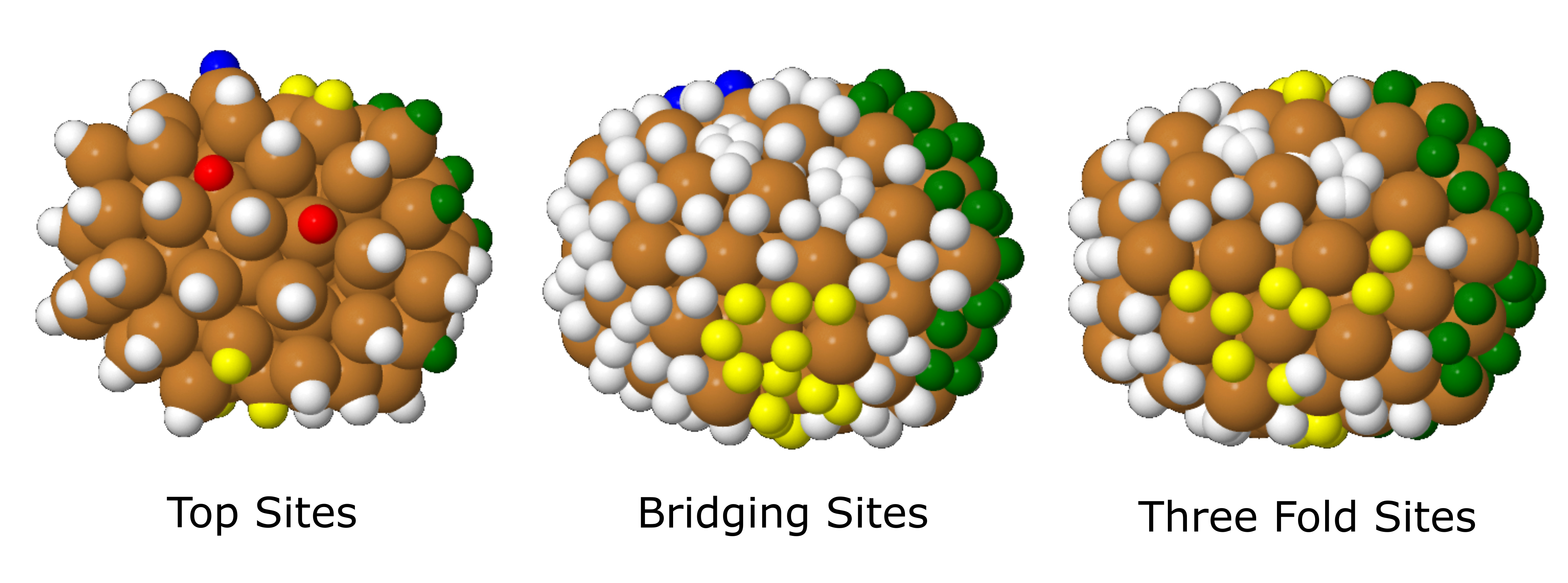
Top sites (SYSTEM_NAME_top_sites.xyz), bridging sites (SYSTEM_NAME_bridging_sites.xyz), and three-fold sites (SYSTEM_NAME_three_fold_sites.xyz) across this cluster, where each of these sites are represented with hydrogen atoms. Colours are used to help record which binding sites have been noted of interest for further optimisation with VASP.
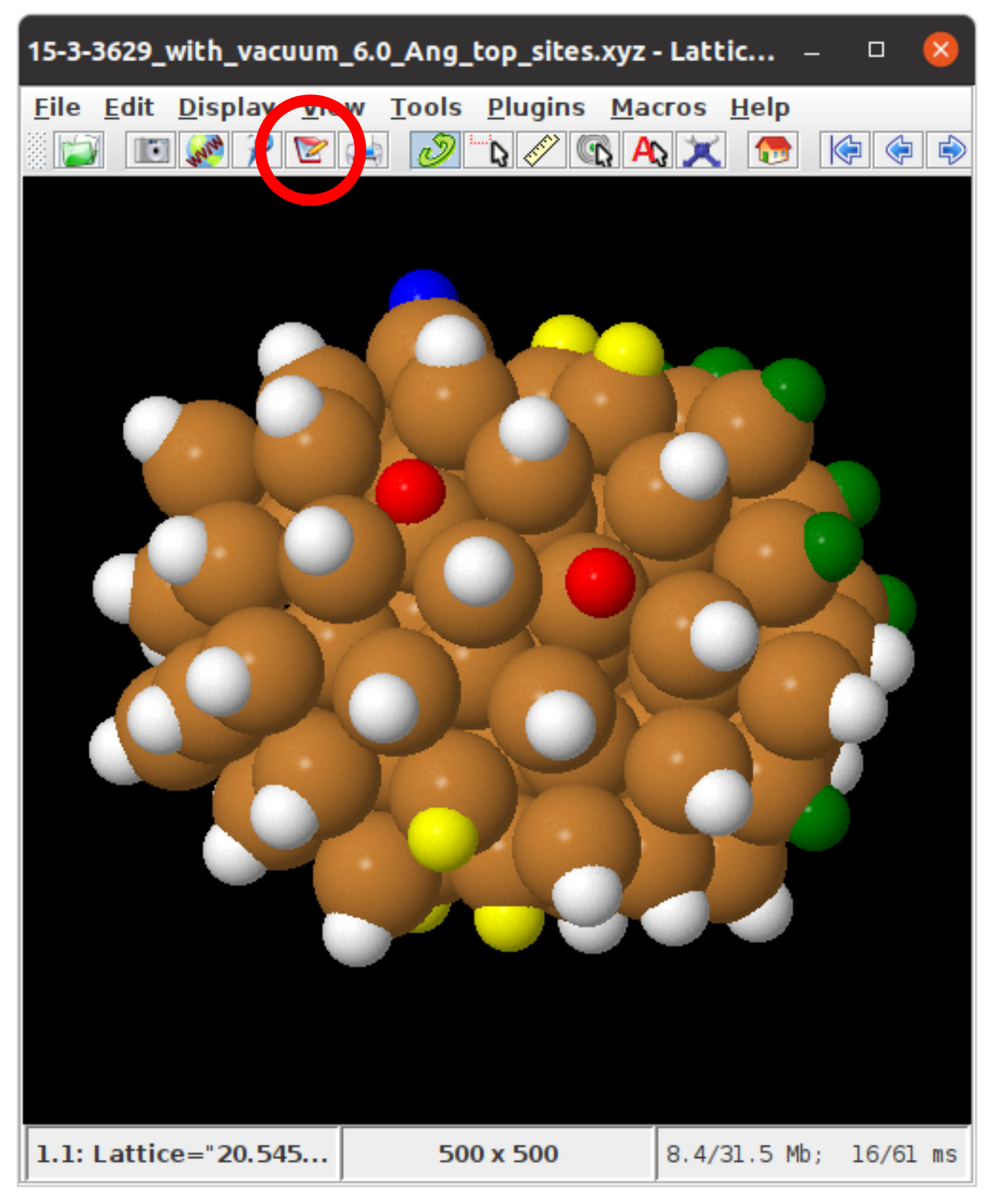
Once you have coloured in your atoms of interest, you can obtain the indices of binding sites of interest by saving your Jmol system as a state file. You can do this by clicking on the notepad icon circled in red in the figure below:
If you open this file in a notepad program (for example in Sublime, see https://www.sublimetext.com/) and scroll down to the section called function _setModelState(), the indices of the atoms your have coloured are given here. For example in the section of the state file shown below:
function _setModelState() {
select ({89 96 98 102 115 121 131 135});
color atoms opaque [xffff00];
select ({0:77});
Spacefill 1.2;
select ({104 126});
color atoms opaque [xff0000];
select ({92 93 132});
color atoms opaque [x0000ff];
select ({78:135});
Spacefill 0.66;
select ({106:109 112 113 118 119 124 130});
color atoms opaque [x008000];
hover "%U";
frank off;
font frank 16.0 SansSerif Plain;
select *;
set fontScaling false;
}
Every select ({Indices}); line that comes before a color atoms line are the indices of the atoms that you have select for binding adsorbates to. You can copy the ADSORBATE_ADSORPTIONSITE_Label_Index.xyz files from your Part_B_All_Systems_with_Adsorbed_Species\ADSORBATE\ADSORPTIONSITE folder to the corresponding Part_C_Selected_Systems_with_Adsorbed_Species_to_Convert_into_VASP_files\ADSORBATE\ADSORPTIONSITE folder by hand. Here, you want to look at the Index of your ADSORBATE_ADSORPTIONSITE_Label_Index.xyz and compare these to your entries in the relevant select ({Indices}); lines.
How to automate the copying of these xyz files: Using copy_files_from_folder_B_to_C.py
The process of choosing which binding sites to use for adsorbating adsorbates to can be a laborious process. For this reason, I have created another python script called copy_files_from_folder_B_to_C.py which can copy the relevant files for you. An example of this is shown below:
from Adsorber import Copy_Files_from_Folder_B_to_Folder_C
adsorbates = ['CO', 'COOH']
top_sites = {'Weird_Sites_Yellow': '89 96 98 102 115 121 131 135', '5_Fold_Vertex_Site_Red':'104 126', 'Weird_Corners_Blue':'92 93 132', 'Ico_Sites_Green':'106:109 112 113 118 119 124 130'}
bridge_sites = {'Weird_Sites_Yellow':'100 109 114 115 119:122 132 135 141:143 148 149 160 171', 'Other_5_fold_Sites_Blue':'99 123 126 127 130 131 150 152 157 158 227 229 241 245', 'Ico_Like_Green':'155 164 173:188 191 193 195 197:204 214 217:223 225 228 235 242 244'}
three_fold_sites = {'Weird_Sites_Yellow':'93 95 97 98 101 102 106 109:111 114:117 120 125 136 137 145 174:176 187', 'Ico_Like_Green':'132 133 138:140 147:163 165:170 178:186 191'}
four_fold_sites = {}
Copy_Files_from_Folder_B_to_Folder_C(adsorbates, top_sites, bridge_sites, three_fold_sites, four_fold_sites)
NOTE: You can copy the indices from the function _setModelState method from the Jmol file (as shown above) and paste them into the dictionaries for top_sites, bridge_sites, three_fold_sites, and four_fold_sites.
What will copy_files_from_folder_B_to_C.py do?
This program will copy of the relevant ADSORBATE_ADSORPTIONSITE_Label_Index.xyz files from your Part_B_All_Systems_with_Adsorbed_Species folders to your Part_C_Selected_Systems_with_Adsorbed_Species_to_Convert_into_VASP_files folder. All orientations/rotations of adsorbates are included, therefore you will need to delete those orientations/rotations you do not want to include. These will be place in folders based on the names you gave the binding sites in the dictionaries. For example, you will find the folders Weird_Sites_Yellow, 5_Fold_Vertex_Site_Red, Weird_Corners_Blue, Ico_Sites_Green in your Top_Sites folder in Part_C_Selected_Systems_with_Adsorbed_Species_to_Convert_into_VASP_files.
What To Do Once You Have Placed Selected xyz Files Into Part_C_Selected_Systems_with_Adsorbed_Species_to_Convert_into_VASP_files
Once you have placed the selected adsorbate+system xyz files into Part_C_Selected_Systems_with_Adsorbed_Species_to_Convert_into_VASP_files of the desired orientations/rotations, you can proceed to Part C (Part C1: Preparing Selected Adsorbed Systems For VASP Optimisation).