Part B1: How to Mark Surface Atoms in your Cluster/Surface model in Adsorber
One of the pieces of information that Adsorber need to know are which atoms on your cluster are the surface atoms. The easiest way to figure out which atoms are surface atoms is to open your cluster/surface model in ASE GUI. An example is given below:
An example cluster when observed in ASE GUI.
Then you want to go to show the indices of the atoms in your cluster/surface model by clicking in the menu View > Show Labels > Atom Index. This will show the indices of atoms in your cluster/surface model in your ASE GUI.
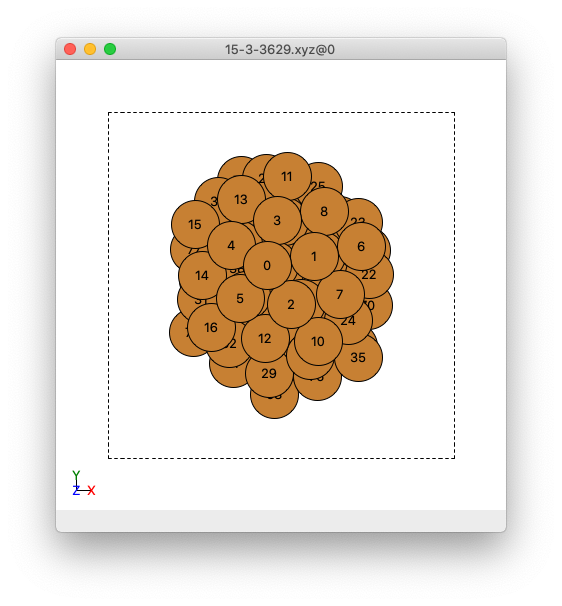
An example cluster when observed in ASE GUI, where atoms have been labelled by their indices.
We will want to include the indices of the surface atoms in your cluster/surface model in the adsorbate.py file in the surface_atoms list. In the example given in the Examples/Cu78_Example/15-3-3629.xyz file, the surface atoms are:
surface_atoms = [11,25,28,13,3,8,6,23,22,59,34,62,66,1,0,4,30,15,14,16,5,12,29,2,7,10,24,26,70,35,47,50,60,63,48,39,41,44,54,68,76,71,32,31,74,42,56,52,43,40,46,61,53,45,57,72,73,77]
The Adsorber program will create a .xyz file called SYSTEM_NAME_tagged_surface_atoms.xyz that will have all surface atoms tagged 1 and all non-surface atoms tagged 0 (where SYSTEM_NAME is the name of the .xyz or .traj file that you gave for the name variable in general.py, see Prelude 1: Setting your general.py and adsorbate.py scripts). You can see this if you open SYSTEM_NAME_tagged_surface_atoms.xyz in ASE GUI, show atom index label by clicking View > Show Labels > Atom Index, and colouring in atoms based on their tag by clicking View > Colors and selecting By tag:
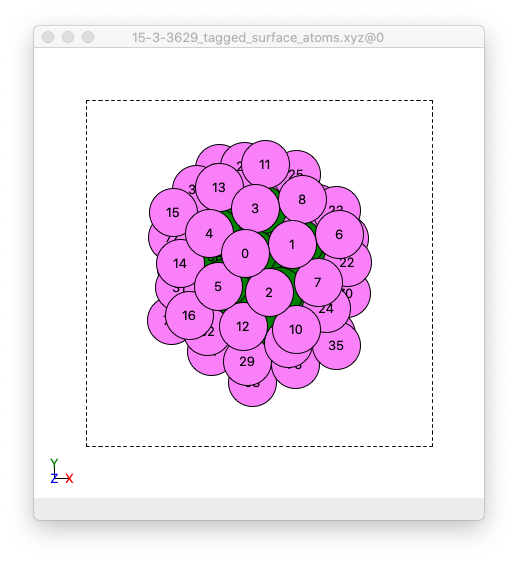
Front view of an image of the example cluster where surface atoms are coloured pick, and non-surface atoms coloured green. This example .xyz file is created by Adsorber.
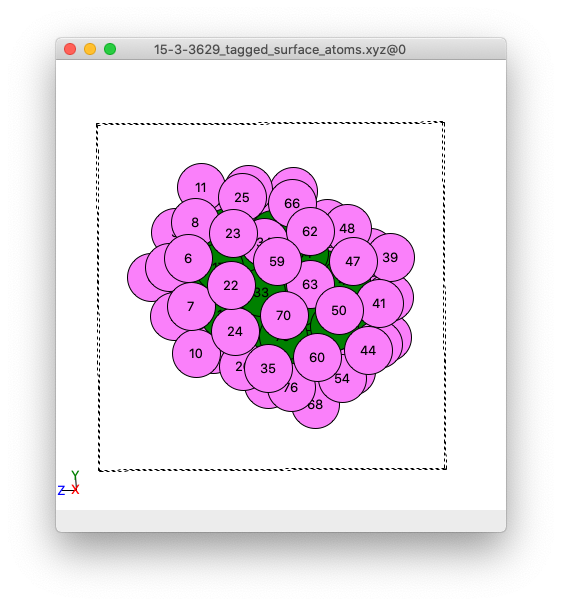
Side view of an image of the example cluster where surface atoms are coloured pick, and non-surface atoms coloured green. This example .xyz file is created by Adsorber.
All the surface atoms should be coloured pink, while the non-surface atoms coloured green. If there are any bulk atom coloured pink or surface atoms coloured green, you will need to remove or add the indices of atoms from the surface_atoms list in the adsorbate.py file to make sure that surface_atoms reflects the atoms in the cluster/surface model that are in fact surface atoms.